|
I am a senior ML Scientist II at Genentech in the Prescient Design team in New York, USA. I am developping ML generative and predictive models for molecular design, more recently focusing on de novo generation. I obtained a PhD degree at Sorbonne Université. My PhD research focused on improving out-of-distribution generalization of deep learning models in the setting of classification and physical dynamics modelling. I obtained a Master's degree at Mines Paris - PSL and at Ecole Normale Supérieure Paris Saclay (Master MVA).Email / Google Scholar / Semantic Scholar / GitHub / LinkedIn / Twitter |

|
|
|
|
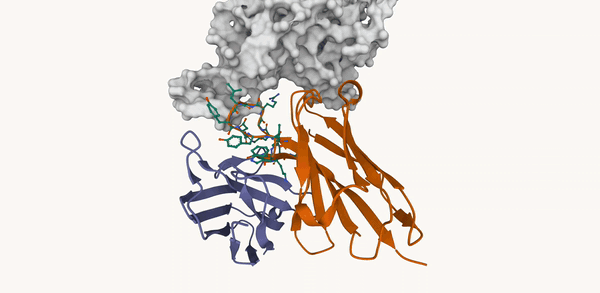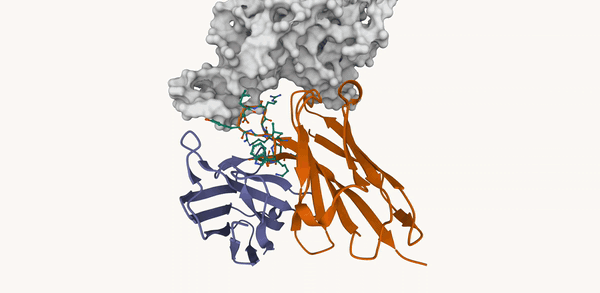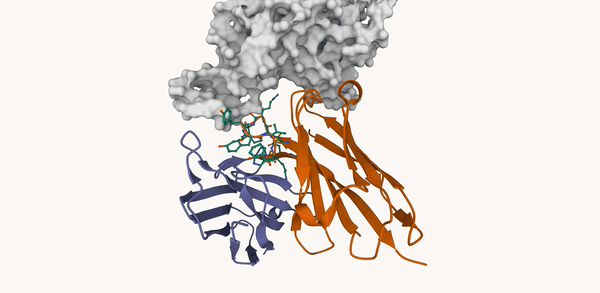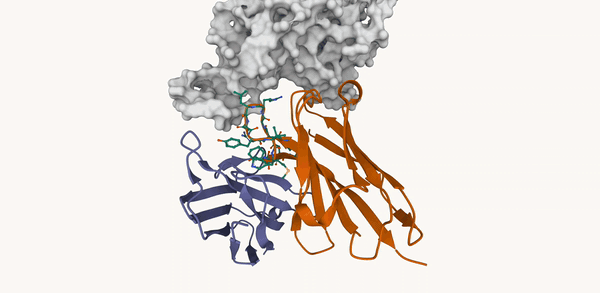
M. Kirchmeyer*, P. O. Pinheiro*, E. Willett, K. Martinkus, J. Kleinhenz, E. K. Makowski, A. M. Watkins, V. Gligorijevic, R. Bonneau, S. Saremi NeurIPS 2025 ArXiv / OpenReview / code |
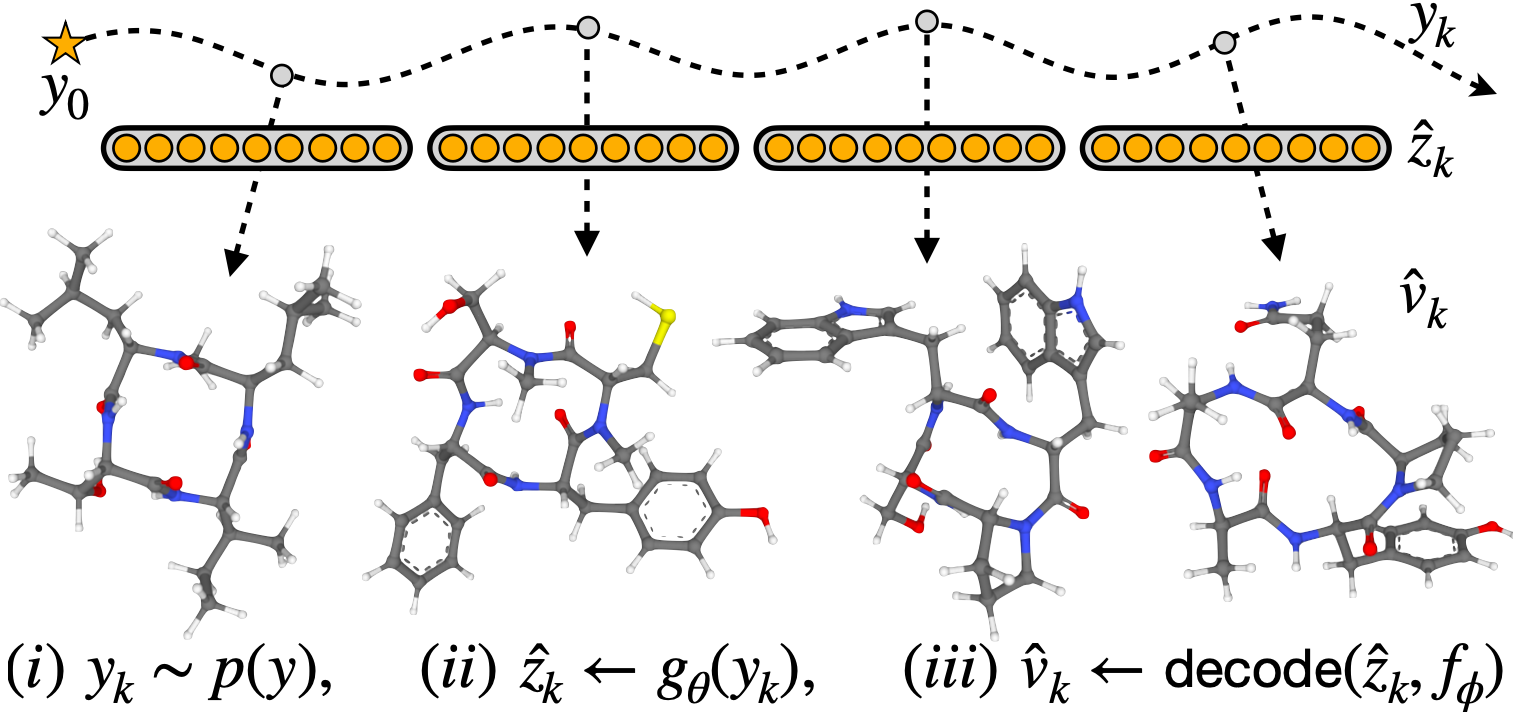
|
M. Kirchmeyer*, P. O. Pinheiro*, S. Saremi NeurIPS 2024 ArXiv / OpenReview / code / |
|
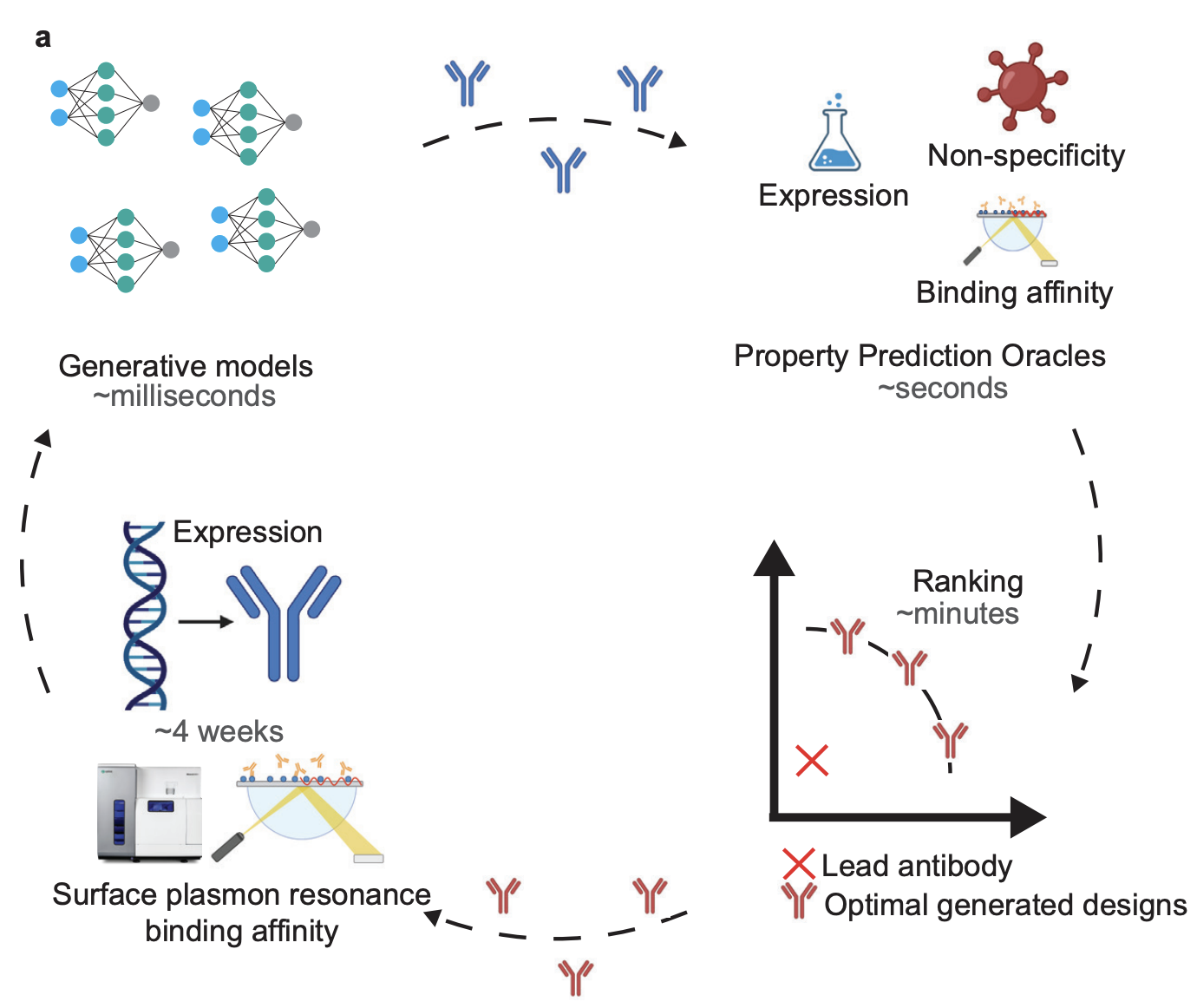
N. Frey*, I. Hötzel*, S. Stanton*, R. Kelly*, R. Alberstein*, ..., M. Kirchmeyer, ..., V. Gligorijevic. BioRxiv ArXiv / |
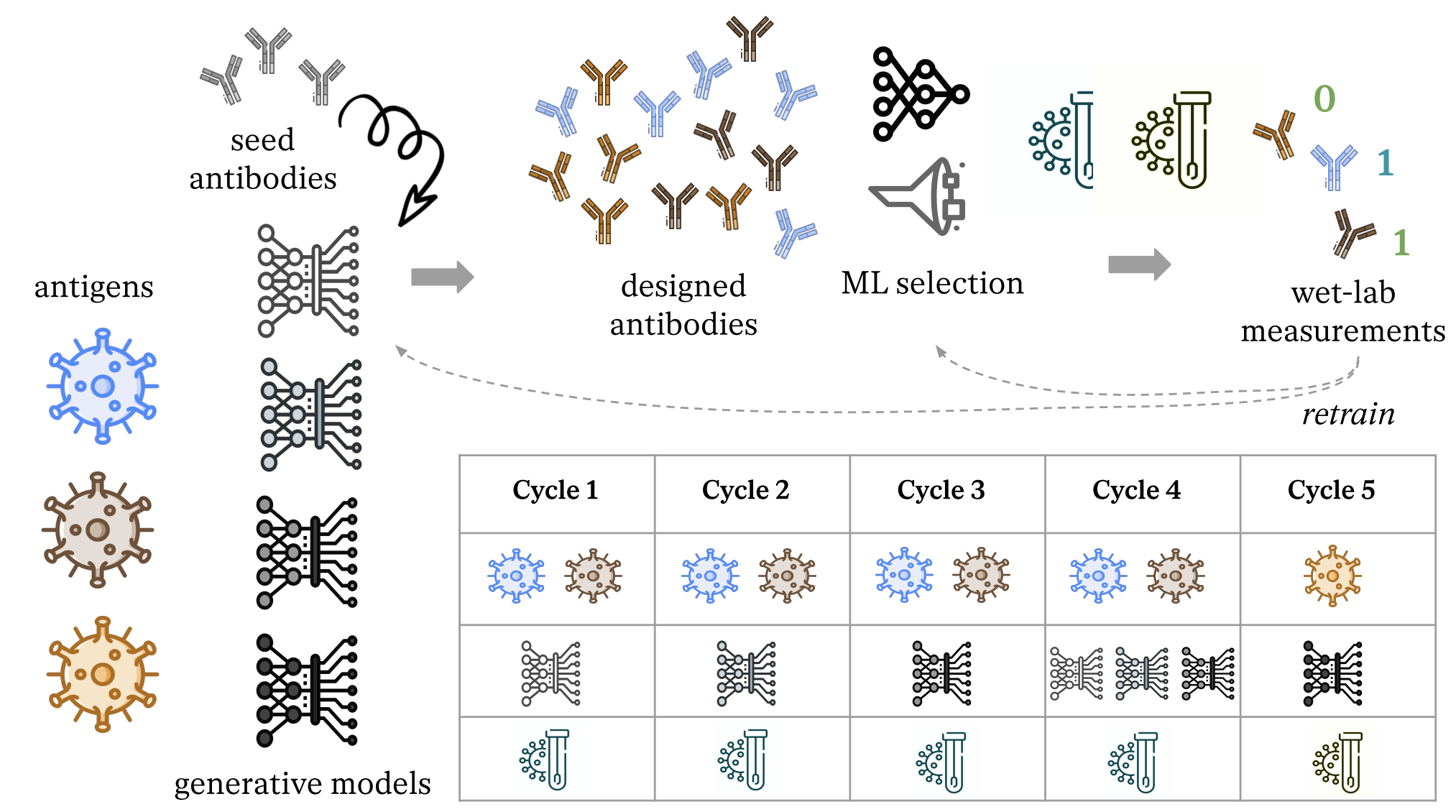
|
N. Tagasovka*, J. Park*, M. Kirchmeyer, ..., K. Cho ICML Workshop on Spurious Correlations, Invariance and Stability, 2023 ArXiv / code / |

|
Y. Yin*, M. Kirchmeyer*, J-Y. Franceschi*, A. Rakotomamonjy, P. Gallinari ICLR 2023 - Spotlight (notable-top-25%) arXiv / OpenReview / code / |
|
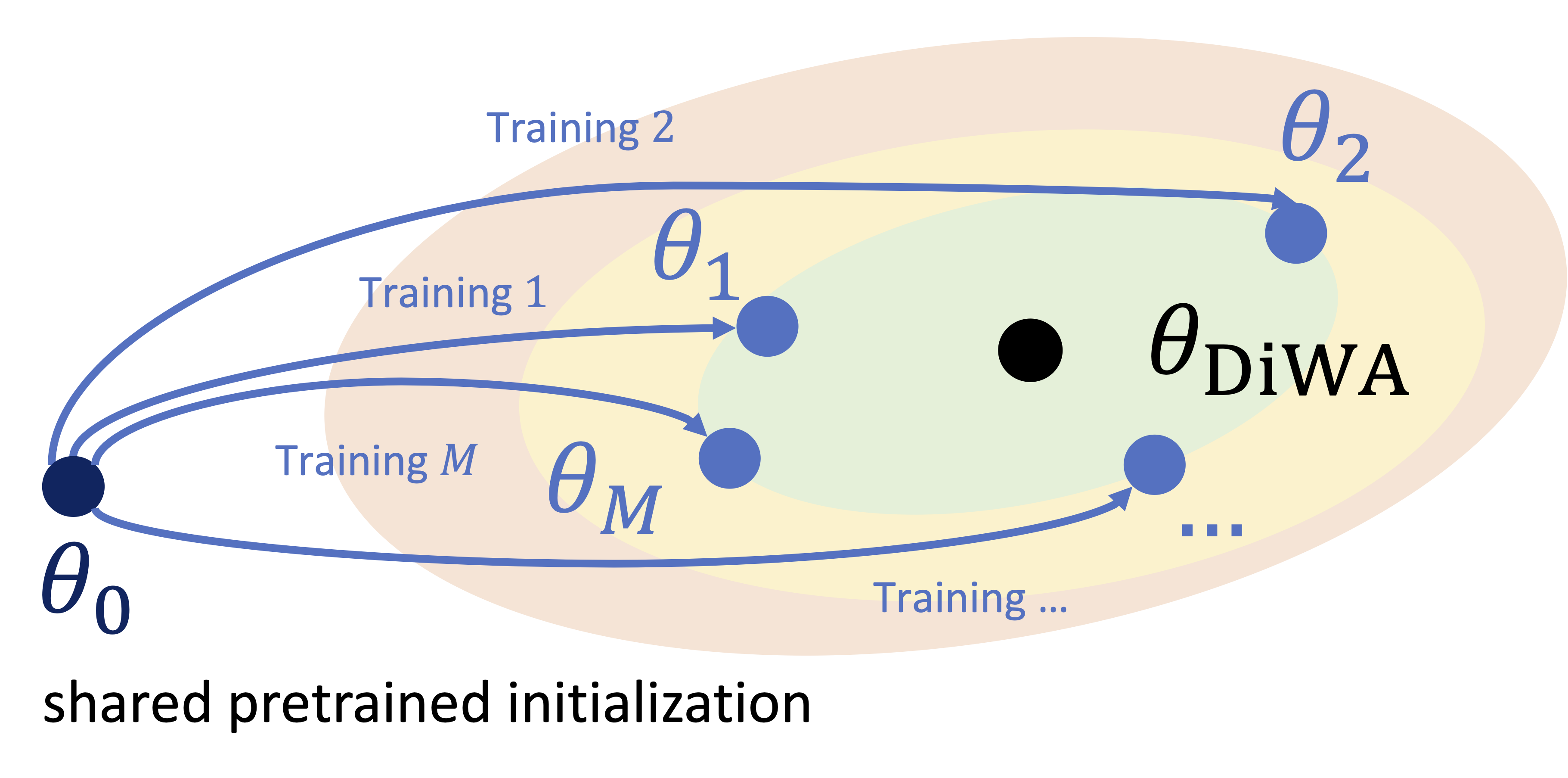
A. Ramé*, M. Kirchmeyer*, T. Rahier, A. Rakotomamonjy, P. Gallinari, M. Cord NeurIPS 2022 arXiv / OpenReview / code / slides / poster |
|
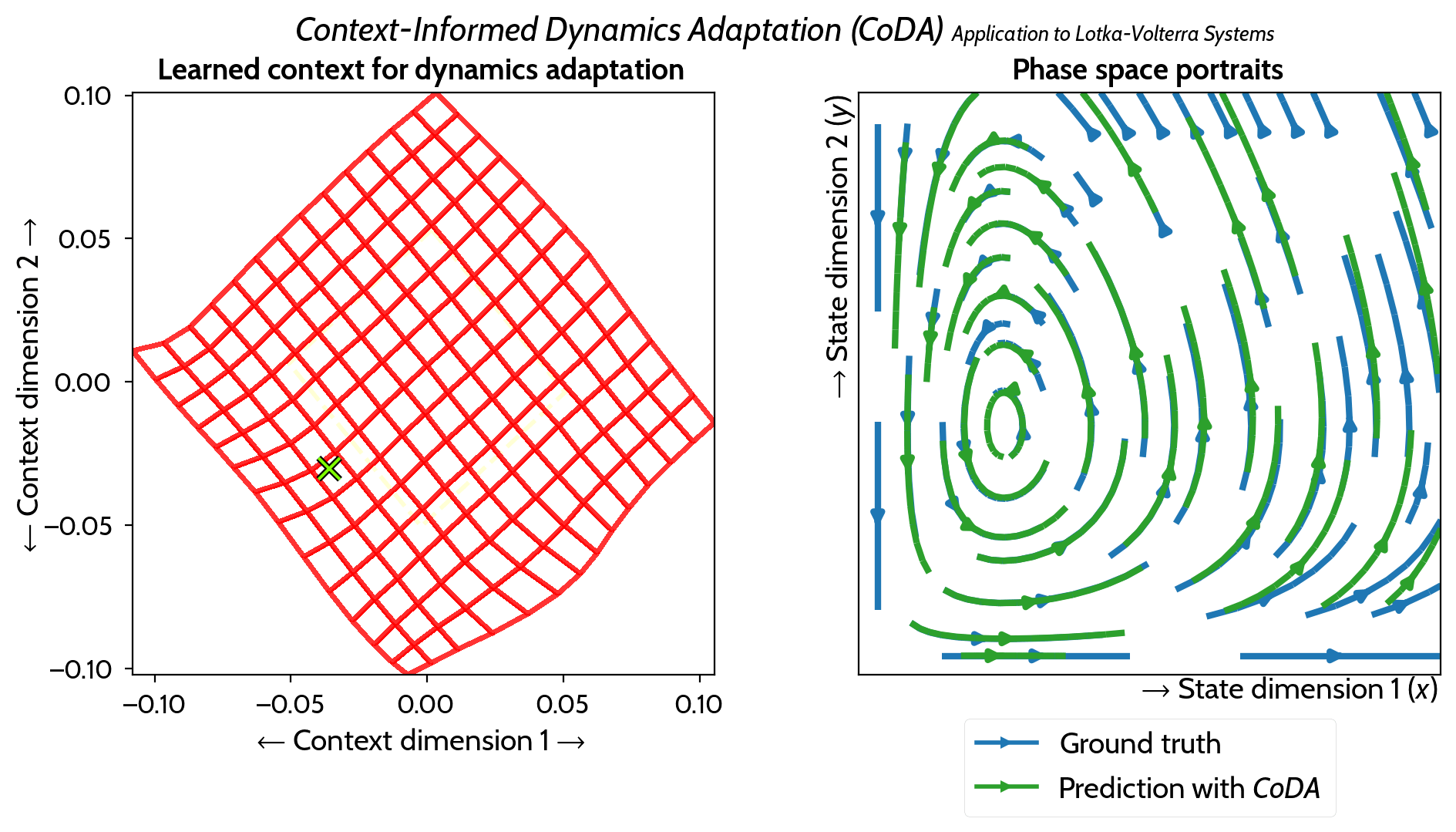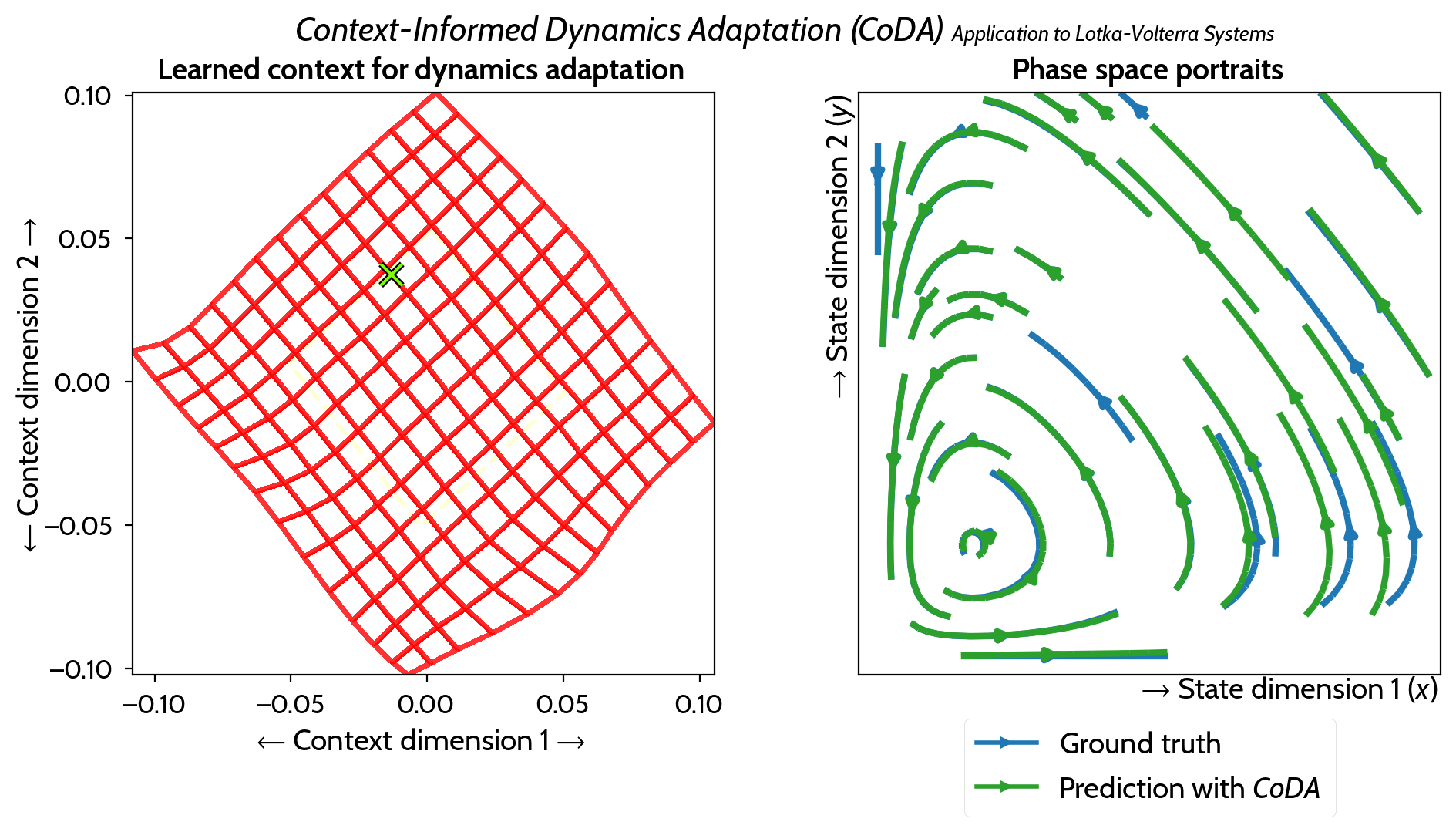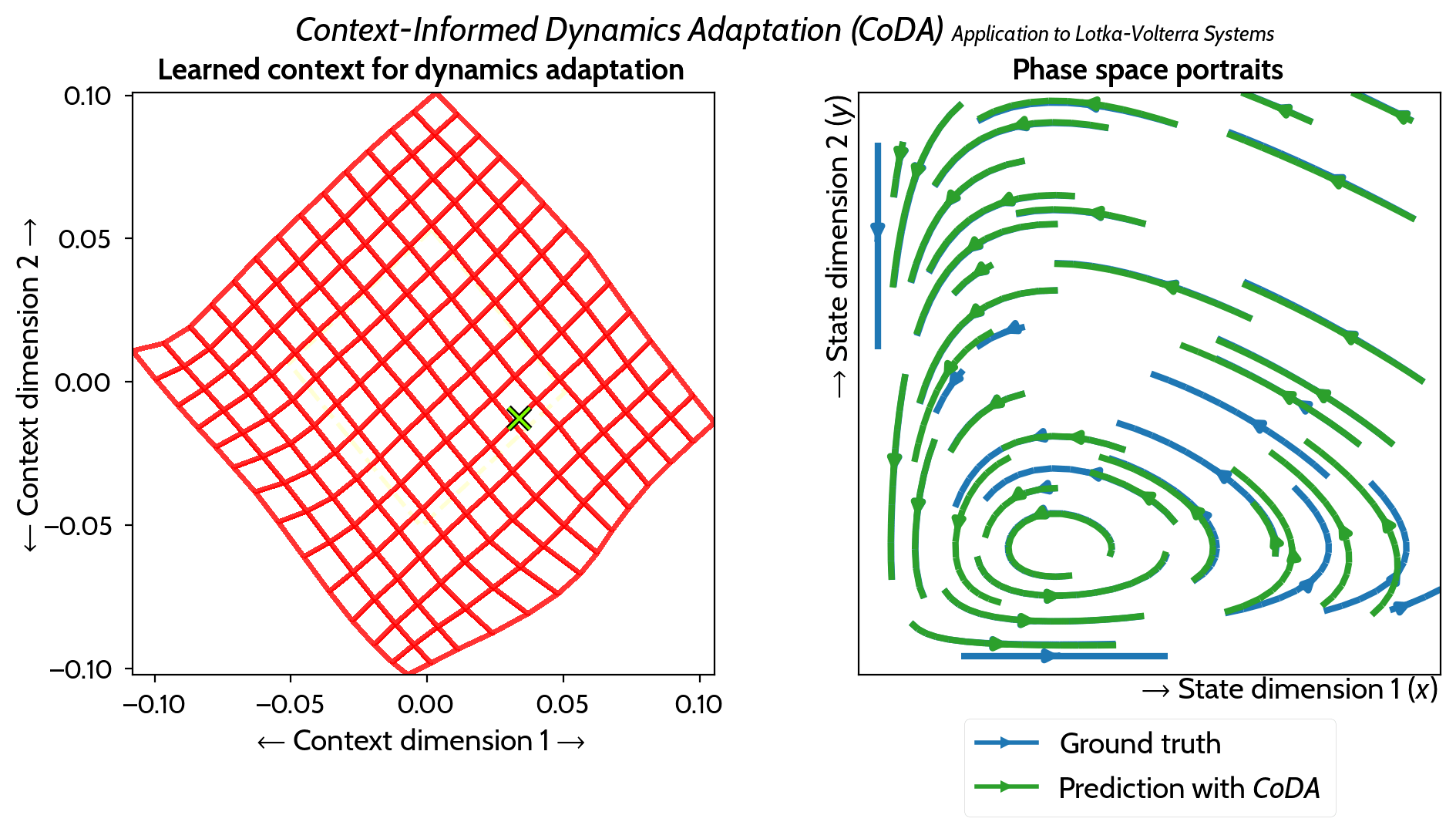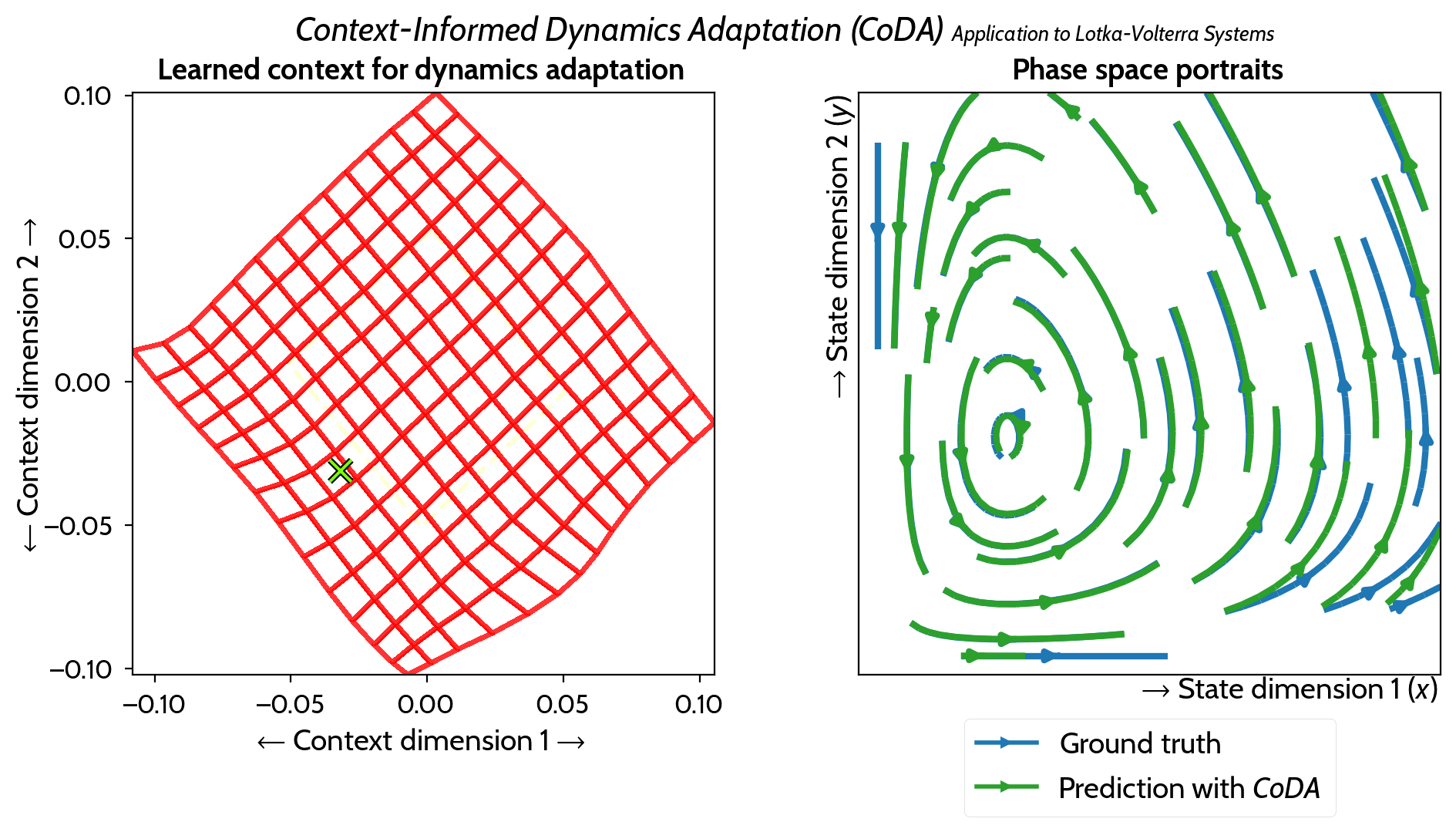
M. Kirchmeyer*, Y. Yin*, J. Donà, N. Baskiotis, A. Rakotomamonjy, P. Gallinari ICML 2022 arXiv / PMLR / code / slides / poster / video |
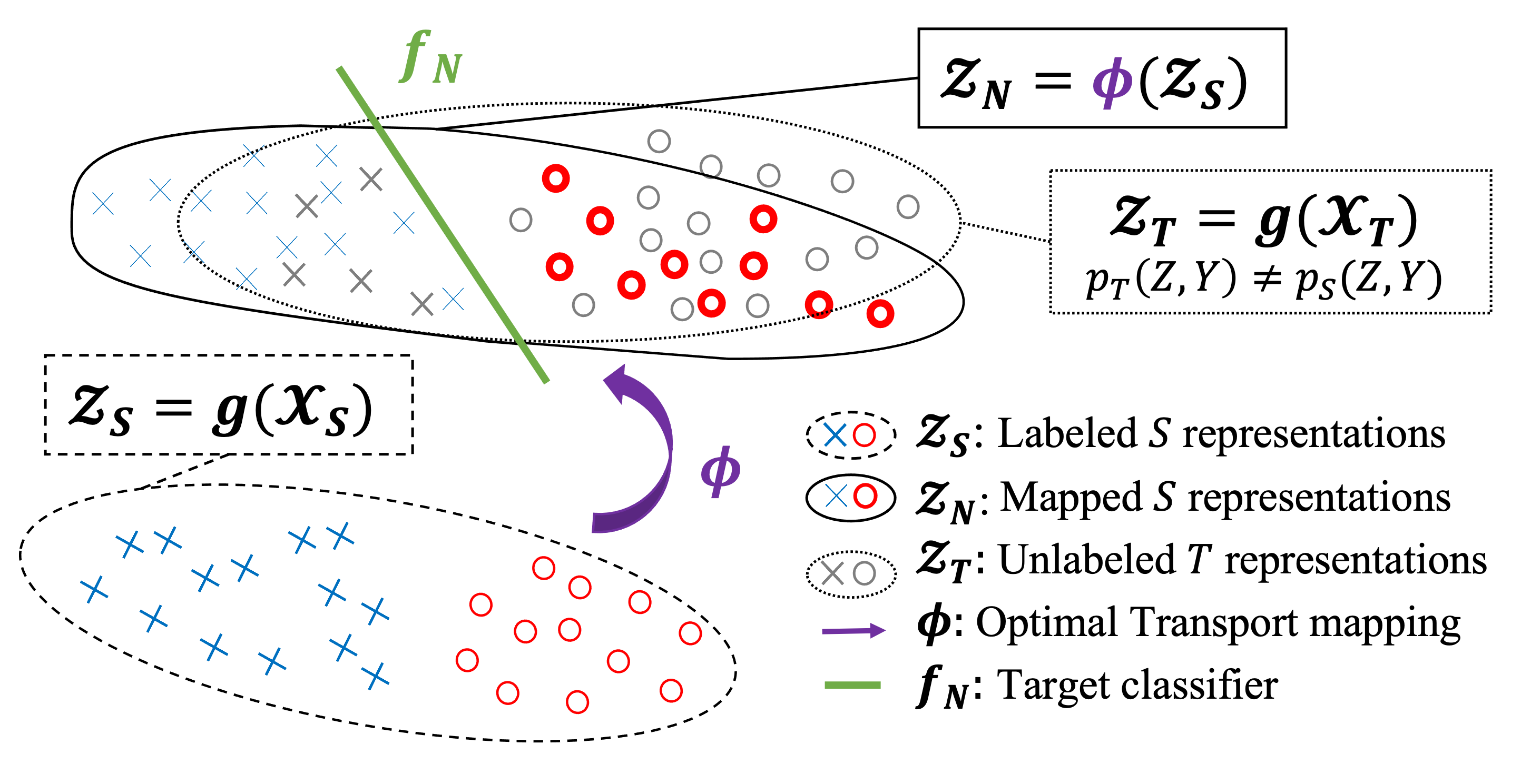
|
M. Kirchmeyer, A. Rakotomamonjy, E. de Bézenac, P. Gallinari ICLR 2022 arXiv / OpenReview / code / slides / poster / video |
|
Template modified from here |